-Search query
-Search result
Showing 1 - 50 of 82 items for (author: torii & s)

EMDB-18942: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - F particle
Method: single particle / : Kumar K, Antanasijevic A

EMDB-18943: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - A particle
Method: single particle / : Kumar K, Antanasijevic A

EMDB-18944: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - E particle
Method: single particle / : Kumar K, Antanasijevic A

PDB-8r5x: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - F particle
Method: single particle / : Kumar K, Antanasijevic A

PDB-8r5y: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - A particle
Method: single particle / : Kumar K, Antanasijevic A

PDB-8r5z: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - E particle
Method: single particle / : Kumar K, Antanasijevic A

EMDB-29745: 
Cryo-EM structure of the Mismatch Sensing Complex (I) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

EMDB-29746: 
Cryo-EM structure of the Mismatch Uncoupling Complex (II) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D
EMDB-29747: 
Cryo-EM structure of the Wedge Alignment Complex (VIII) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D
EMDB-29748: 
Cryo-EM structure of the Primer Separation Complex (IX) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

EMDB-29749: 
Cryo-EM structure of the Mismatch Locking Complex (III) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

EMDB-29750: 
Cryo-EM structure of the Guide loop Engagement Complex (VI) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

EMDB-29751: 
Cryo-EM structure of the Guide loop Engagement Complex (IV) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

EMDB-29752: 
Cryo-EM structure of the Guide loop Engagement Complex (V) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D
EMDB-41091: 
Cryo-EM structure of the Backtracking Initiation Complex (VII) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

PDB-8g5i: 
Cryo-EM structure of the Mismatch Sensing Complex (I) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

PDB-8g5j: 
Cryo-EM structure of the Mismatch Uncoupling Complex (II) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

PDB-8g5k: 
Cryo-EM structure of the Wedge Alignment Complex (VIII) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

PDB-8g5l: 
Cryo-EM structure of the Primer Separation Complex (IX) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

PDB-8g5m: 
Cryo-EM structure of the Mismatch Locking Complex (III) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

PDB-8g5n: 
Cryo-EM structure of the Guide loop Engagement Complex (VI) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

PDB-8g5o: 
Cryo-EM structure of the Guide loop Engagement Complex (IV) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

PDB-8g5p: 
Cryo-EM structure of the Guide loop Engagement Complex (V) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D

PDB-8t7e: 
Cryo-EM structure of the Backtracking Initiation Complex (VII) of Human Mitochondrial DNA Polymerase Gamma
Method: single particle / : Nayak AR, Buchel G, Herbine KH, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D
EMDB-33972: 
Cryo-EM structure of the SARS-CoV-2 spike protein (3-up RBD) bound to neutralizing antibody CSW1-1805
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y
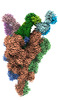
EMDB-33973: 
Cryo-EM structure of the SARS-CoV-2 spike protein (1-up RBD) bound to neutralizing antibody CSW2-1353
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y
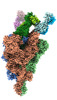
EMDB-33974: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing antibody CSW2-1353
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y
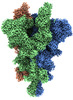
EMDB-33975: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) with C480A mutation
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y

EMDB-16246: 
ARE-ABCF VmlR2 bound to a 70S ribosome
Method: single particle / : Crowe-McAuliffe C, Wilson DN

PDB-8buu: 
ARE-ABCF VmlR2 bound to a 70S ribosome
Method: single particle / : Crowe-McAuliffe C, Wilson DN

EMDB-13242: 
PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state I
Method: single particle / : Crowe-McAuliffe C, Wilson DN

PDB-7p7r: 
PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state I
Method: single particle / : Crowe-McAuliffe C, Wilson DN

EMDB-13241: 
E. faecalis 70S ribosome bound by PoxtA-EQ2, high-resolution combined volume
Method: single particle / : Crowe-McAuliffe C, Wilson DN

EMDB-13243: 
PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state II
Method: single particle / : Crowe-McAuliffe C, Wilson DN

EMDB-13244: 
PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state III
Method: single particle / : Crowe-McAuliffe C, Wilson DN

EMDB-13245: 
E. faecalis 70S ribosome with P-tRNA, state IV
Method: single particle / : Crowe-McAuliffe C, Wilson DN

PDB-7p7q: 
E. faecalis 70S ribosome bound by PoxtA-EQ2, high-resolution combined volume
Method: single particle / : Crowe-McAuliffe C, Wilson DN

PDB-7p7s: 
PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state II
Method: single particle / : Crowe-McAuliffe C, Wilson DN

PDB-7p7t: 
PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state III
Method: single particle / : Crowe-McAuliffe C, Wilson DN

PDB-7p7u: 
E. faecalis 70S ribosome with P-tRNA, state IV
Method: single particle / : Crowe-McAuliffe C, Wilson DN

EMDB-13017: 
RqcH DR variant bound to 50S-peptidyl-tRNA-RqcP RQC complex (rigid body refinement)
Method: single particle / : Crowe-McAuliffe C, Wilson DN

PDB-7ope: 
RqcH DR variant bound to 50S-peptidyl-tRNA-RqcP RQC complex (rigid body refinement)
Method: single particle / : Crowe-McAuliffe C, Wilson DN

EMDB-30915: 
Apo spike protein of SARS-CoV2
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30916: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody 8D2
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30917: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (8D2)-1
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30918: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (8D2)-2
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30919: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (8D2)-3
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30920: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (8D2)-4
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30921: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (2940)-1
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30922: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (2940)-2
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
